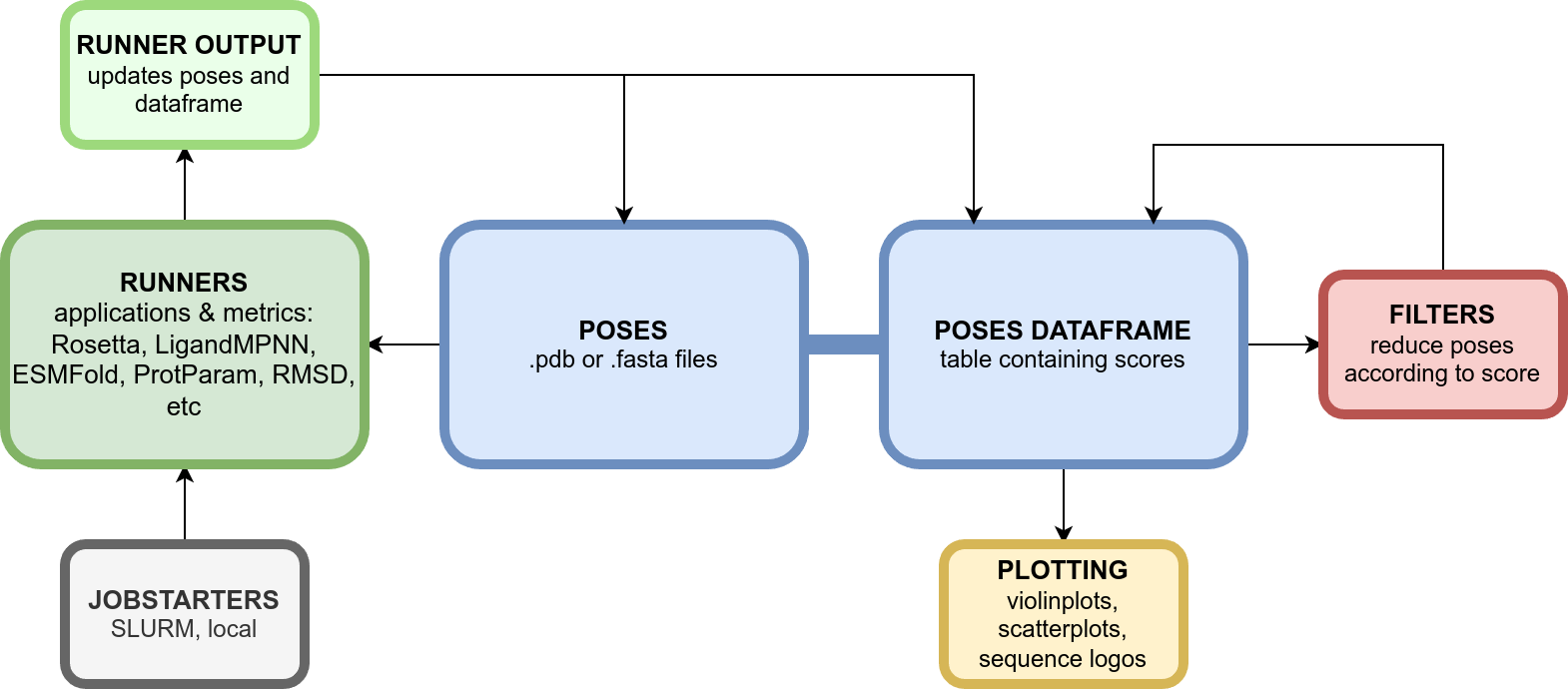
A Python package for running protein design tools on SLURM clusters.
First, clone the repository
git clone https://github.com/mabr3112/ProtFlow.git
ProtFlow requires python >= 3.11. You can either install it into an existing environment, or you can create a new one with like this:
conda create -n protflow python=3.11
conda activate protflow
Install the package in development mode (we are currently under active development).
cd ProtFlow
pip install -e .
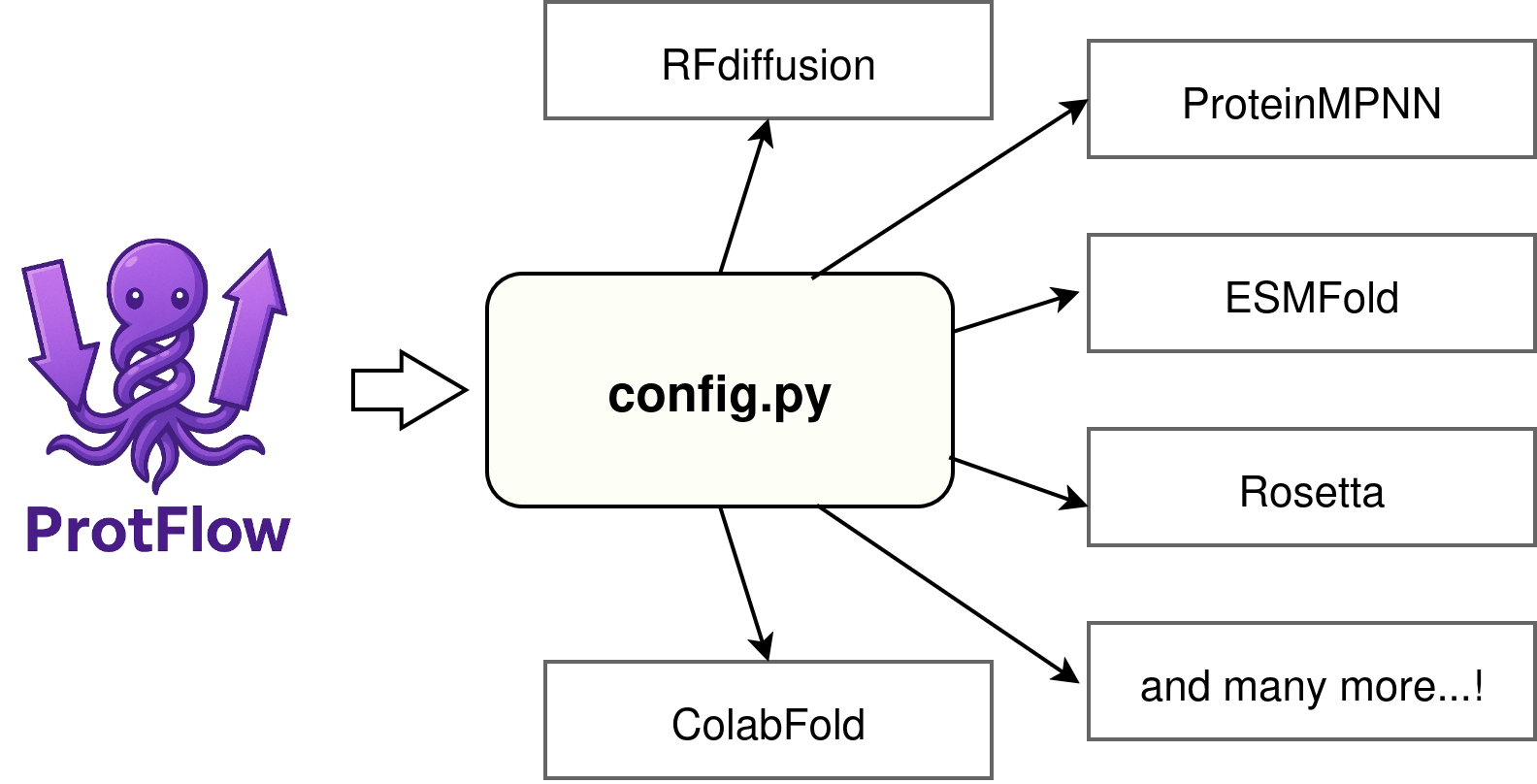
After the install, instantiate your config.py file. This can be done with the command below, which will copy the config.py file from protflow/config_template.py
protflow-init-config
By default protflow-init-config will set up config.py in ~/.config/protflow/config.py.
If you want to link your protflow installation to a different config.py file elswhere, there is a simple cli-tool, protflow-set-config.
Linking your protflow to a different config.py might come in handy if you want to share it system wide on a computing cluster with multiple users.
Simply provide the path to the target config.py to the tool:
protflow-set-config /path/to/your/config.py
If you want to unset this custom override, supply the --unset flag:
protflow-set-config --unset
Finally, to check which config.py file your protflow is currently using:
protflow-check-config
ProtFlow is documented with read-the-docs and sphinx. The documentation can be found here: https://protflow.readthedocs.io/en/latest/