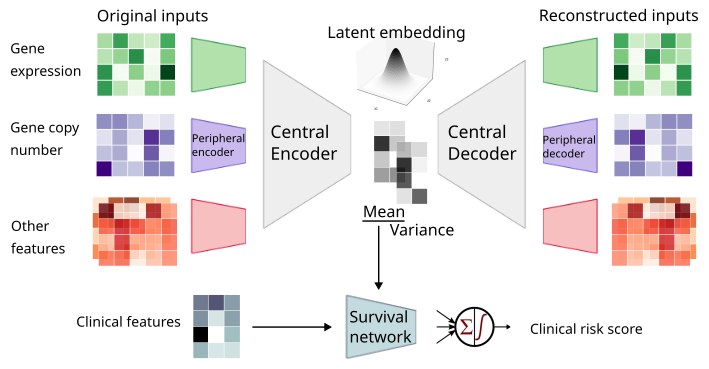
MyeVAE is a variational autoencoder leveraging multi-omics for risk prediction in newly diagnosed multiple myeloma patients.
This repository contains the Python source code to preprocess multimoodal features, train MyeVAE, score its performance, and perform SHAP analysis.
All computation is done on the CPU, using the PyTorch library.
Requires Python 3.9 or later
Install through PyPI
pip install myevaeInstall through github
pip install git+https://github.com/JiaGengChang/myevae.gitDownload the raw multi-omics dataset and example outputs from the following link:
-
Input datasets are stored at https://myevae.s3.us-east-1.amazonaws.com/example_inputs.tar.gz
-
Example output files are stored at https://myevae.s3.us-east-1.amazonaws.com/example_outputs.tar.gz
Please use the download links sparingly.
If you use the AWS S3 CLI, the bucket ARN is arn:aws:s3:::myevae - you can list its contents and download specific files.
As there are over 50,000 raw features and contains missing values, this step performs supervised-feature selection, scaling, and imputation.
For feature selection, elastic net regularized Cox proportional hazards is used.
For imputation, round-robin imputation with scikit-learn IterativeImputer is used, with random forest regressor and classifier as based estimators.
myevae preprocess \
-e [endpoint: os | pfs] \
-i [/inputs/folder]The following output files will be created in the same directory as the inputs.
project
└──inputs
├──...
├──train_features_os_processed.csv
├──train_features_pfs_processed.csv
├──valid_features_os_processed.csv
└──valid_features_pfs_processed.csvPreprocessing of features as computationally expensive than hyperparameter tuning. On 10 cores and 64GB RAM, it uses a wall time of ~12 hours.
Alternatively, if you are just testing this package, you may use pre-processed features from the S3 bucket link and proceed to the next step.
See requirements for the required files before training.
Basically, you need to place the processed features and labels in /inputs/folder, and create a hyperparameter file named param_grid.py in /params/folder. See this section for the default hyperparamter grid.
myevae train \
-e [endpoint: os | pfs] \
-n [model_name] \
-i [/inputs/folder] \
-p [/params/folder] \
-t [nthreads] \
-o [/output/folder]If the hyperparameter file is not specified, the default will be used (/src/myevae/param_grid.py), the process should take only 5 minutes.
With an actual hyperparameter grid, the training can take up to 12 hours.
myevae score \
-e [endpoint: os | pfs] \
-n [model_name] \
-i [/inputs/folder] \
-o [/output/folder]Model scoring should be done in under a minute.
myevae shap \
-e [endpoint: os | pfs] \
-n [model_name] \
-i [/inputs/folder] \
-o [/output/folder]Approximation of SHAP values and generation of summary plots should take 1-2 minutes.
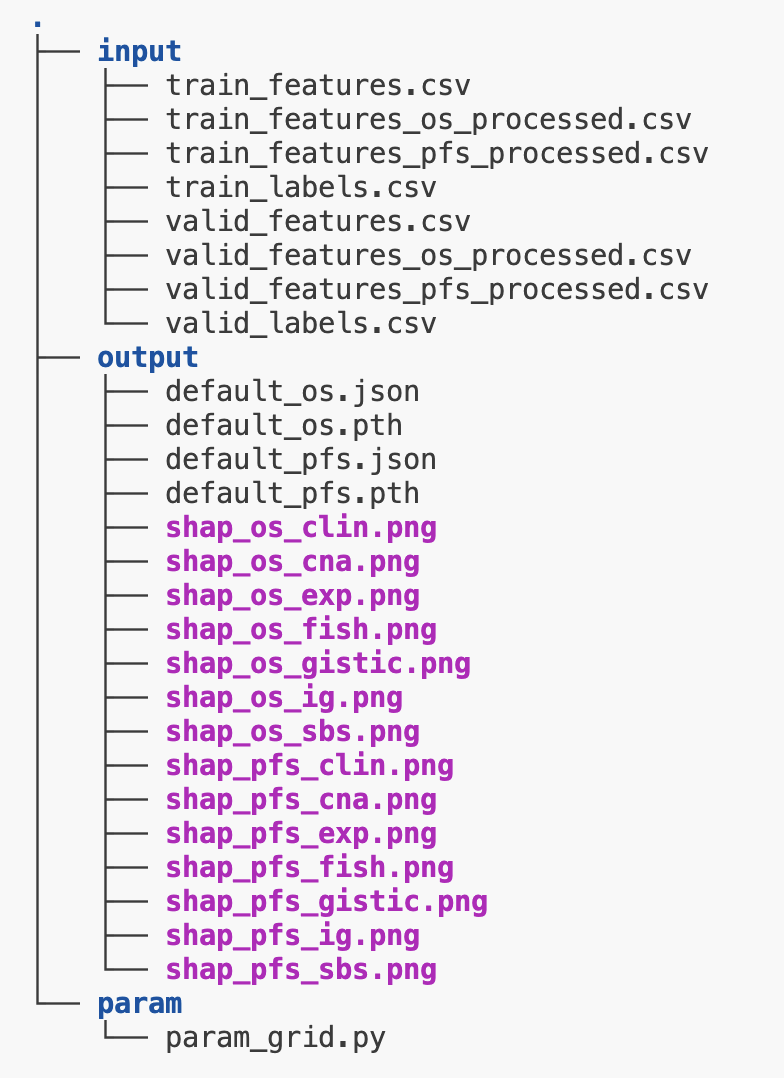
For a model named "default", the final output should look something like this.
Validation scores and best parameters can be found in the .json files. The .pth files are model weights.
Required for preprocessing only.
Place the following .csv files in the inputs folder:
project
└──inputs
├──train_features_os_processed.csv
├──valid_features_os_processed.csv
└──train_labels.csvNote the preprocessing does not use valid labels (valid_labels.csv), but it is fine if the file is in the directory.
Required for training, scoring and shap analysis
Place the following .csv files inside your inputs folder (/inputs/folder), and the param_grid.py inside your preferred folder. Also create an empty folder for the outputs.
project
├──inputs
│ ├──train_features_os_processed.csv
│ ├──valid_features_os_processed.csv
│ ├──train_labels.csv
│ └──valid_labels.csv
├──params
│ └──param_grid.py
└──outputs
└──[output files created here]For features*.csv and labels*.csv, column 0 is read in as the index, which should be the patient IDs.
Example input csv files can be downloaded from AWS S3 link above.
Required for training only.
Place a python file named param_grid.py containing the hyperparameter grid dictionary in the params folder (specified with -p).
This contains the set of hyperparameters that grid search will be performed on.
Otherwise, use the default provided in src/myevae/param_grid.py. This default hyperparameter grid is only meant for testing purposes:
from torch.nn import LeakyReLU, Tanh
# the default hyperparameter grid for debugging uses
# this is not meant to be used for real training, as the search space is only on z_dim
param_grid = {
'z_dim': [8,16,32],
'lr': [5e-4],
'batch_size': [1024],
'input_types': [['exp','cna','gistic','fish','sbs','ig']],
'input_types_subtask': [['clin']],
'layer_dims': [[[32, 4],[16,4],[4,1],[4,1],[4,1],[4,1]]],
'layer_dims_subtask' : [[4,1]],
'kl_weight': [1],
'activation': [LeakyReLU()],
'subtask_activation': [Tanh()],
'epochs': [100],
'burn_in': [20],
'patience': [5],
'dropout': [0.3],
'dropout_subtask': [0.3]
}An actual paramater file is available at https://myevae.s3.us-east-1.amazonaws.com/param_grid.py
Using the actual hyperparameter file, the training time will be significantly longer (~12 hours)
These dependencies will be automatically installed.
python >= 3.9
torch >= 1.9.0
scikit-learn >= 0.24.1
scikit-survival >= 0.23.1
importlib_resources
matplotlib
shap-0.47.3.dev8-offline-fork-for-myevae==0.0.1
The last dependency is a offline fork of shap (https://pypi.org/project/shap/) which has been modified to work MyeVAE.
-
Minimum 4 CPU cores (8 cores is recommended)
-
Minimum 16 GB RAM (64GB is required for feature preprocessing)
No GPU is required.
If you use MyeVAE in your research, please consider citing:
Jia Geng Chang, Jianbin Chen, Guo-Liang Chew, Wee Joo Chng. MyeVAE: a multi-modal variational autoencoder for risk profiling of newly diagnosed multiple myeloma. 2 May 2025. Manuscript under review.