Description
I am posting a picture of the pluto notebook for better understanding/readability (what are the outputs etc). Below you can find copyable code.
Copyable Code
begin
using Flux:Flux
using MLDatasets: MNIST
using LinearAlgebra: norm, LinearAlgebra
end
MNIST.download(i_accept_the_terms_of_use=true)
begin
x_train, y_train = MNIST.traindata()
x_train = Float32.(x_train)
y_train_oh = Flux.onehotbatch(y_train, 0:9)
size(x_train), size(y_train), size(y_train_oh)
end
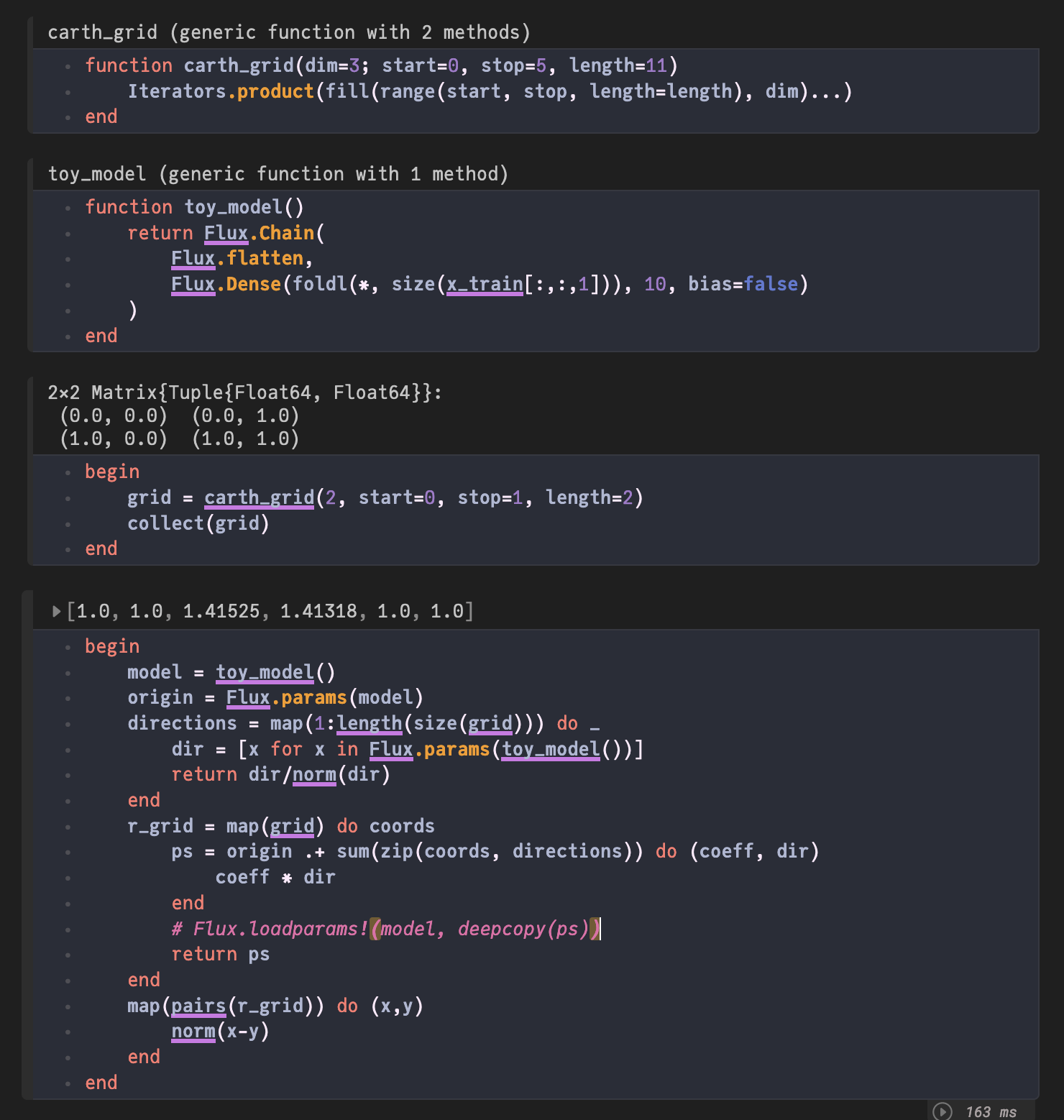
function carth_grid(dim=3; start=0, stop=5, length=11)
Iterators.product(fill(range(start, stop, length=length), dim)...)
end
function toy_model()
return Flux.Chain(
Flux.flatten,
Flux.Dense(foldl(*, size(x_train[:,:,1])), 10, bias=false)
)
end
begin
grid = carth_grid(2, start=0, stop=1, length=2)
collect(grid)
end
begin
model = toy_model()
origin = Flux.params(model)
directions = map(1:length(size(grid))) do _
dir = [x for x in Flux.params(toy_model())]
return dir/norm(dir)
end
r_grid = map(grid) do coords
ps = origin .+ sum(zip(coords, directions)) do (coeff, dir)
coeff * dir
end
# Flux.loadparams!(model, deepcopy(ps))
return ps
end
map(pairs(r_grid)) do (x,y)
norm(x-y)
end
endOkay so what is happening? I define a generator of a model toy_model, to essentially generate different parameter initializations. Because apparently that is faster than using loadparams #1764 and I can use the default initializer to get a more realistic distribution over initializations. Then I sample two prameter vectors from this distribution and normalize them. Since they are random with high dimension, they are very likely to be almost orthogonal. So due to normalization we get orthonormal vectors. So when I create the grid using the coordinates from a carthesian grid, I am essentially doing an orthonormal basis change.
Okay, so far so good. Now when I determine the distances of all the points on the new grid, the end result should be the same as before the basis change. Now the longest distance is between the points (0,0) and (1,1). Which is the squareroot of 2. So ~1.4. And that is in fact the largets value of the output. Nice! Everything works as intended.
But if I comment in the Flux.loadparams! my new output makes no sense
[1.0, 1.41621, 2.83242, 1.0, 2.23859, 1.41621]
This implies that loadparams! somehow modifies ps even through the deepcopy?